 Your shopping cart is currently empty
Your shopping cart is currently empty

KU-55933 (ATM Kinase Inhibitor) is an ATM inhibitor (IC50=12.9 nM; Ki=2.2 nM) that is selective and ATP-competitive. KU-55933 induces apoptosis and has antitumor effects.
| Pack Size | Price | USA Warehouse | Global Warehouse | Quantity |
|---|---|---|---|---|
| 5 mg | $43 | In Stock | In Stock | |
| 10 mg | $61 | In Stock | In Stock | |
| 25 mg | $103 | In Stock | In Stock | |
| 50 mg | $166 | In Stock | In Stock | |
| 100 mg | $288 | In Stock | - | |
| 1 mL x 10 mM (in DMSO) | $48 | In Stock | In Stock |
| Description | KU-55933 (ATM Kinase Inhibitor) is an ATM inhibitor (IC50=12.9 nM; Ki=2.2 nM) that is selective and ATP-competitive. KU-55933 induces apoptosis and has antitumor effects. |
| Targets&IC50 | ATM:12.9 nM, DNA-PK:2500 nM, PI3K:16600 nM, mTOR:9300 nM |
| In vitro | METHODS: Ovarian cancer cells SKOV-3 were treated with Eflornithine hydrochloride hydrate (1-100 µM) for 48 h. Cell viability was measured by PrestoBlue assay. RESULTS: Eflornithine significantly inhibited the viability of SKOV-3 cells in a dose-dependent manner. [1] METHODS: MYCN2 (-) and MYCN2 (+) NB cells were treated with Eflornithine hydrochloride hydrate (5 mM) and putrescine/spermidine/spermine for 72 h. Cell migration was detected by wound healing assay. RESULTS: Significant differences in cell migration were observed between untreated control cells and Eflornithine-treated cells. eflornithine inhibited cell migration by 73% and 72% in MYCN2 (-) and MYCN2 (+) cells, respectively. Supplementation of the medium with polyamines attenuated the effect of Eflornithine on MYCN2 (-) and MYCN2 (+) cells. [2] |
| In vivo | METHODS: To test the anti-infective activity in vivo, Eflornithine hydrochloride hydrate (25-50 mg/kg) and hydroxyurea (50-100 mg/kg) were orally administered to B. microti infected BALB/c mice once daily for 5 days. RESULTS: Oral administration of hydroxyurea and Eflornithine inhibited the proliferation of Bifidobacterium microti in mice by 60.1% and 78.2%, respectively. hydroxyurea-DA and Eflornithine-DA combination treatment showed higher efficacy of chemotherapy than monotherapy. [3] |
| Kinase Assay | Purified enzyme assays: ATM for use in the in vitro assay is obtained from HeLa nuclear extract by immunoprecipitation with rabbit polyclonal antiserum raised to the COOH-terminal 400 amino acids of ATM in buffer containing 25 mM HEPES (pH 7.4), 2 mM MgCl2, 250 mM KCl, 500 μM EDTA, 100 μM Na3VO4, 10% v/v glycerol, and 0.1% v/v Igepal. ATM-antibody complexes are isolated from nuclear extract by incubating with protein A-Sepharose beads for 1 hour and then through centrifugation to recover the beads. In the well of a 96-well plate, ATM-containing Sepharose beads are incubated with 1 μg of substrate glutathione S-transferase–p53N66 (NH2-terminal 66 amino acids of p53 fused to glutathione S-transferase) in ATM assay buffer [25 mM HEPES (pH 7.4), 75 mM NaCl, 3 mM MgCl2, 2 mM MnCl2, 50 μM Na3VO4, 500 μM DTT, and 5% v/v glycerol] at 37 °C in the presence or absence of inhibitor. After 10 minutes with gentle shaking, ATP is added to a final concentration of 50 μM and the reaction continued at 37 °C for an additional 1 hour. The plate is centrifuged at 250 × g for 10 minutes (4 °C) to remove the ATM-containing beads, and the supernatant is removed and transferred to a white opaque 96-well plate and incubated at room temperature for 1.5 hours to allow glutathione S-transferase-p53N66 binding. This plate is then washed with PBS, blotted dry, and analyzed by a standard ELISA technique with a phospho-serine 15 p53 antibody. The detection of phosphorylated glutathione S-transferase-p53N66 substrate is performed in combination with a goat antimouse horseradish peroxidase-conjugated secondary antibody. Enhanced chemiluminescence solution is used to produce a signal and chemiluminescent detection is carried out. |
| Cell Research | U2OS cells are exposed to ionizing radiation (3, 5, or 15 Gy) or UV (5 or 50 J/m2) and the ATM response determined by Western blot analysis of p53 serine 15 phosphorylation and stabilization of wild-type p53. Whole cell extracts are obtained from each time point, proteins separated by SDS-PAGE, and the ATM-specific increase in phosphorylated serine 15 measured with a p53 phospho-serine 15 specific antibody. Overall p53 stabilization with time is also observed with a p53-specific antibody (DO-1). Similarly, for studying ATM-dependent phosphorylations on H2AX, CHK1, NBS1, and SMC1, the following antibodies are used: CHK1 phospho-serine 345 and NBS1 phospho-serine 343 antibodies. Histone H2A (H-124) and CHK1 antibodies are also used, as well as SMC1 and SMC1 phospho-serine 966 antibodies. For determination of a cellular IC50 for KU-55933, the peak response time for p53 serine 15 phosphorylation of 2 hours is used to monitor inhibition of ATM. KU-55933 is titrated onto cells and preincubated for 1 hour before ionizing radiation. Using scanning densitometry, the percentage inhibition relative to vehicle control is calculated, and the IC50 value is calculated as for the in vitro determinations.(Only for Reference) |
| Synonyms | ATM Kinase Inhibitor |
| Molecular Weight | 395.49 |
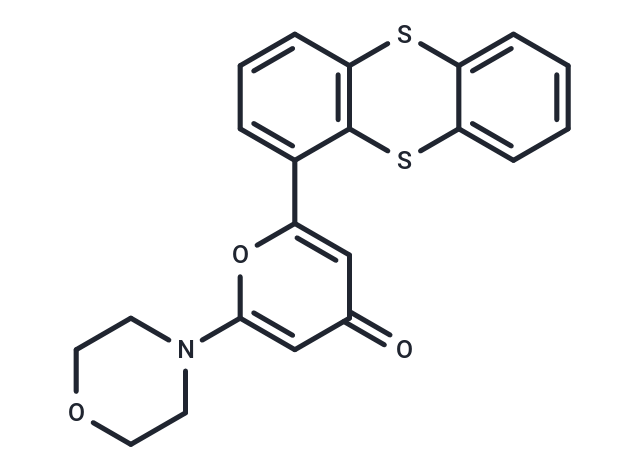
| Formula | C21H17NO3S2 |
| Cas No. | 587871-26-9 |
| Smiles | O=c1cc(oc(c1)-c1cccc2Sc3ccccc3Sc12)N1CCOCC1 |
| Relative Density. | 1.419 g/cm3 (Predicted) |
| Storage | store at low temperature,keep away from moisture | In solvent: -80°C for 1 year | Shipping with blue ice/Shipping at ambient temperature. | |||||||||||||||||||||||||||||||||||
| Solubility Information | Ethanol: 19.8 mg/mL (50.06 mM), Sonication is recommended. DMSO: 16.67 mg/mL (42.15 mM), Sonication is recommended. | |||||||||||||||||||||||||||||||||||
| In Vivo Formulation | 10% DMSO+40% PEG300+5% Tween 80+45% Saline: 2 mg/mL (5.06 mM), Sonication is recommended. Please add the solvents sequentially, clarifying the solution as much as possible before adding the next one. Dissolve by heating and/or sonication if necessary. Working solution is recommended to be prepared and used immediately. The formulation provided above is for reference purposes only. In vivo formulations may vary and should be modified based on specific experimental conditions. | |||||||||||||||||||||||||||||||||||
Solution Preparation Table | ||||||||||||||||||||||||||||||||||||
DMSO/Ethanol
Ethanol
| ||||||||||||||||||||||||||||||||||||
Dissolve 2 mg of the compound in 100 μL DMSO![]() to obtain a stock solution at a concentration of 20 mg/mL . If the required concentration exceeds the compound's known solubility, please contact us for technical support before proceeding.
to obtain a stock solution at a concentration of 20 mg/mL . If the required concentration exceeds the compound's known solubility, please contact us for technical support before proceeding.
1) Add 100 μL of the DMSO![]() stock solution to 400 μL PEG300
stock solution to 400 μL PEG300![]() and mix thoroughly until the solution becomes clear.
and mix thoroughly until the solution becomes clear.
2) Add 50 μL Tween 80 and mix well until fully clarified.
3) Add 450 μL Saline,PBS or ddH2O![]() and mix thoroughly until a homogeneous solution is obtained.
and mix thoroughly until a homogeneous solution is obtained.
| Size | Quantity | Unit Price | Amount | Operation |
|---|

Copyright © 2015-2026 TargetMol Chemicals Inc. All Rights Reserved.